
When human radiologists examine scans, they peer through the lens of decades of training. Extending from college to medical school to residency, the road that concludes in a physician interpreting, say, an X-ray, includes thousands upon thousands of hours of education, both academic and practical, from studying for licensing exams to spending years as a resident.
At present, the training pathway for artificial intelligence (AI) to interpret medical images is much more straightforward: show the AI medical images labeled with features of interest, like cancerous lesions, in large enough quantities for the system to identify patterns that allow it to “see” those features in unlabeled images.
Despite more than 14,000 academic papers having been published on AI and radiology in the last decade, the results are middling at best. In 2018, researchers at Stanford realized that an AI they trained to identify skin lesions erroneously flagged images that contained rulers because most of the images of malignant lesions also had rulers in them.
“Neural networks easily overfit on spurious correlations,” says Mark Yatskar, Assistant Professor in Computer and Information Science (CIS), referring to the AI architecture that emulates biological neurons and powers tools as varied as ChatGPT and image-recognition software. “Instead of how a human makes the decisions, it will take shortcuts.”
Doctors spend years in medical school learning from textbooks and in classrooms before they begin their clinical training in earnest. We’re trying to mirror that process.Mark Yatskar, Assistant Professor in CIS
In a new paper, to be shared at NeurIPS 2024 as a spotlight, Yatskar, together with Chris Callison-Burch, Professor in CIS, and first author Yue Yang, a doctoral student advised by Callison-Burch and Yatskar, introduces a novel means of developing neural networks for medical image recognition by emulating the training pathway of human physicians. “Generally, with AI systems, the procedure is to throw a lot of data at the AI system, and it figures it out,” says Yatskar. “This is actually very unlike how humans learn — a physician has a multi-step process for their education.”
The team’s new method effectively takes AI to medical school by providing a set body of medical knowledge culled from textbooks, from PubMed, the academic database of the National Library of Medicine, and from StatPearls, an online company that provides practice exam questions for medical practitioners. “Doctors spend years in medical school learning from textbooks and in classrooms before they begin their clinical training in earnest,” points out Yatskar. “We’re trying to mirror that process.”
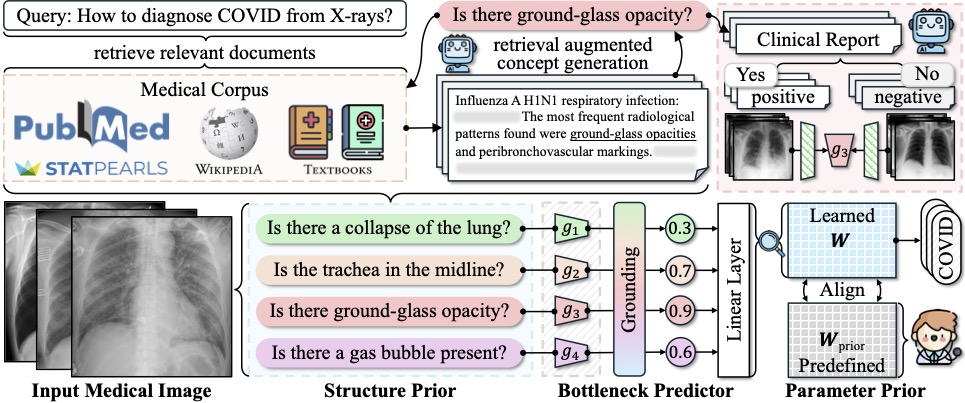
The new approach, deemed Knowledge-enhanced Bottlenecks (KnoBo), essentially requires that AI base decisions on established medical knowledge. “When reading an X-ray, medical students and doctors ask, is the lung clear, is the heart a normal size,” Yang says. “The model will rely on similar factors to the ones humans use when making a decision.”
The upshot is that models trained using KnoBo are not only more accurate at tasks like identifying COVID patients based on lung X-rays than the current best-in-class models, they are also more interpretable: clinicians can understand why the model made a particular decision. “You will know why the system predicts this X-ray is a COVID patient — because it has opacity in the lung,” says Yang.
Models trained with KnoBo are also more robust and able to handle some of the messiness of real-world data. One of the greatest assets of human doctors is that you can place them in many different contexts — with different hospitals and different patient populations — and expect their skills to transfer. In contrast, AI systems trained on a particular group of patients from a particular hospital rarely work well in different contexts.

To assess the ability of KnoBo to help models focus on salient information, the researchers tested a wide range of neural networks on “confounded” data sets, in essence, training the models on one set of patients, where, say, all sick patients were white and healthy patients Black, and then testing the models on patients with the opposite characteristics. “The previous methods fail catastrophically,” says Yang. “Using our way, we constrain the model to reasoning over those knowledge priors we learn from medical documents.” Even on confounded data, models trained using KnoBo averaged 32.4% greater accuracy than neural networks finetuned on medical images.
Given that the American Association of Medical Colleges (AAMC) projects a shortage of 80,000 physicians in the United States alone by 2036, the researchers hope their work will open the door to the safe application of AI in medicine. “You could really make an impact in terms of getting people help that otherwise they couldn’t get because there aren’t people appropriately qualified to give that help,” says Yatskar.
Additional co-authors include Mona Gandhi of The Ohio State University, Yufei Wang of Penn Engineering, Yifan Wu of Meta AI, and Michael S. Yao and Professor James C. Gee of Penn Medicine.
This study was conducted at the University of Pennsylvania School of Engineering and Applied Science and supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via the HIATUS Program contract #2022-22072200005. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for governmental purposes, notwithstanding any copyright annotation therein.
Michael S. Yao was supported by the National Institutes of Health (F30 MD020264). James C. Gee was also supported by the NIH (R01 EB031722).
